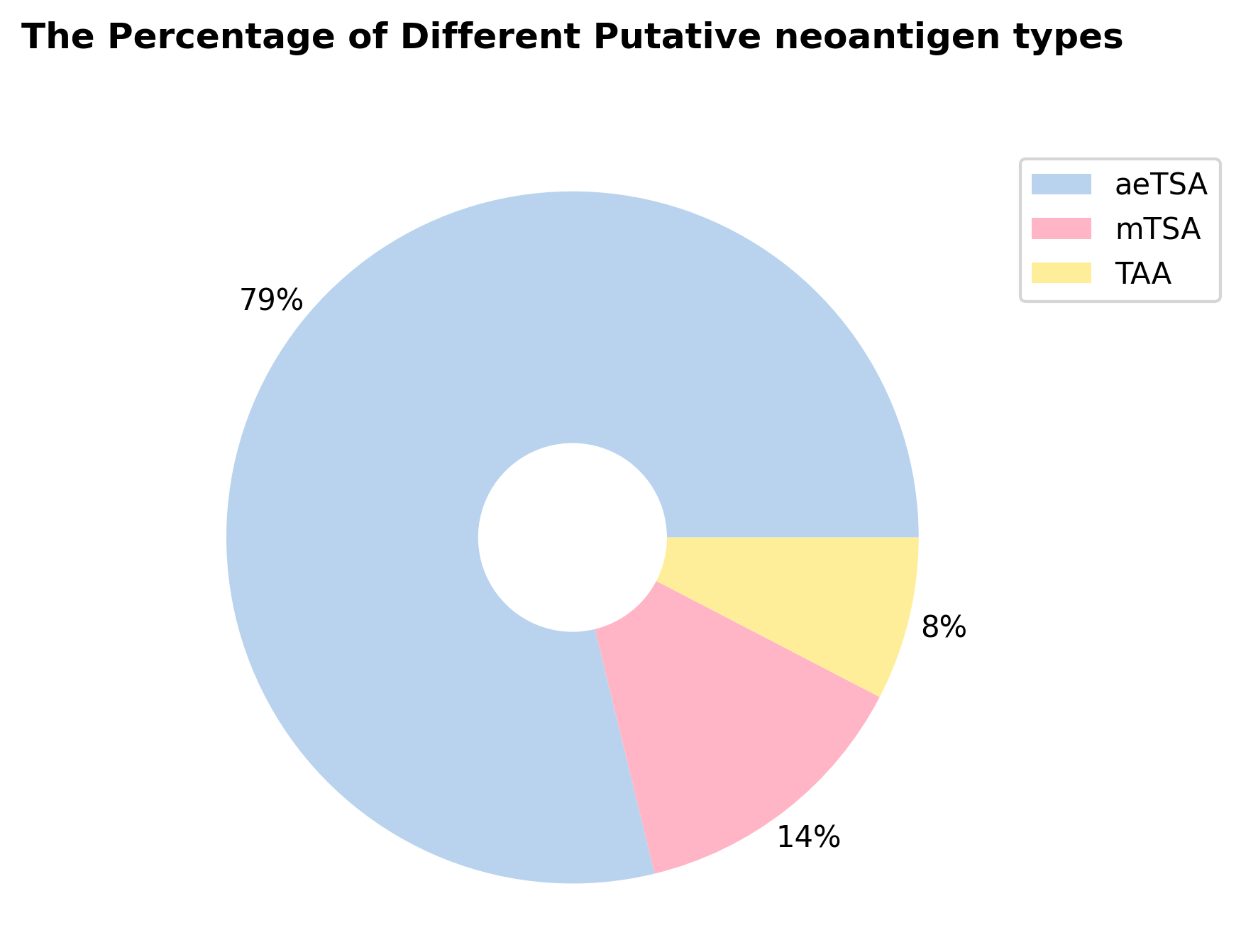NARWHAL
NARWHAL — Neoantigens Recognition Website and HLA Genotyping Tool
Start Neoantigen Start HLA typing Overview
Overview
Congradulations! Your job has been completed successfully!
The followings are details about your job and logs/results. Please click the left navigation panel to show results of each step.
Information about your job:
| Job ID | f8a97207312df3edde7e788440366c32fbd17985af8344f86c94d8ab7d9f9c96 |
| Submission time | May 1, 2024, 8:52 a.m. |
| Start time | May 1, 2024, 8:52 a.m. |
| End time | May 1, 2024, 8:55 p.m. |
| Download the mTSA & aeTSA candidate file (TSV format) | Download |
 Quality Control
Quality Control
Quality Control (QC) Result
The following results include FastQC files and MultiQC files for both tumor and normal samples.
Summary
| DNA-seq Before Trimming |
| DNA-seq After Trimming |
| RNA-seq Before Trimming |
| RNA-seq After Trimming |
DNA-seq
DNA-seq QC report links: Tumor R1
| FastQC before trimming | raw reads | ||
| FastQC after trimming | paired reads | unpaired reads |
DNA-seq QC report links: Tumor R2
| FastQC before trimming | raw reads | ||
| FastQC after trimming | paired reads | unpaired reads |
DNA-seq QC report links: Normal R1
| FastQC before trimming | raw reads | ||
| FastQC after trimming | paired reads | unpaired reads |
DNA-seq QC report links: Normal R2
| FastQC before trimming | raw reads | ||
| FastQC after trimming | paired reads | unpaired reads |
Tumor Trimmomatic
| log file |
Normal Trimmomatic
| log file |
RNA-seq
RNA-seq QC report links: Tumor R1
| FastQC before trimming | raw reads | |
| FastQC after trimming | paired reads |
RNA-seq QC report links: Tumor R2
| FastQC before trimming | raw reads | |
| FastQC after trimming | paired reads |
RNA-seq QC report links: Normal R1
| FastQC before trimming | raw reads | |
| FastQC after trimming | paired reads |
QC report links: Normal R2
| FastQC before trimming | raw reads | |
| FastQC after trimming | paired reads |
Tumor Trimmomatic
| log file |
Normal Trimmomatic
| log file |
 Somatic Mutation Calling
Somatic Mutation Calling
Somatic Mutation Result
The following results include: (1) a compressed VCF file generated by GATK Mutects, (2) a VCF file obtained from GATK SelectVariants, and (3) a TSV file containing contamination segments.
Mutect2
| T.somatic.mutation.vcf.gz |
Variant Selection
 |
T.somatic.variant.vcf |
Contamination Calculation
| T.segment.statistic.tsv |
 Germline Mutation Calling
Germline Mutation Calling
Germline Mutation Result
The following results include: (1) a compressed VCF file generated by GATK HaplotypeCaller and (2) a VCF file obtained from GATK SelectVariants.
 Genome Phasing and Annotation
Genome Phasing and Annotation
Genome Phasing and Annotation Result
The following results include somatic and phased variants' (1) summary reports of VEP annotation and (2) annotated variant files in VCF format.
Annotated Somatic Mutations
| T.somatic.summary.html | T.somatic.vcf |
Annotated Phased Mutations
| T.phased.summary.html | T.phased.vcf |
 RNA Expression Level Calculation and Filtering
RNA Expression Level Calculation and Filtering
RNA Expression Level Result
The following results include: (1) RNA expression levels of all genes in tumor, and (2) RNA expression levels of all genes in normal.
 mTSA Identification with DNA-seq & aeTSA Identification with RNA-seq
mTSA Identification with DNA-seq & aeTSA Identification with RNA-seq
TSA Identification Report
The following files represent mTSA (from DNA-seq) and aeTSA (from RNA-seq) reports under various filtered conditions.
Total TSA Result (in TSV format): Download
mTSA Result (in TSV format): Download
mTSA Result (in FASTA format): Download
aeTSA Result (in TSV format): Download
aeTSA Result (in FASTA format): Download